Family Markers: Using Multiply-Affected Families to Identify Risk Genes
Overview
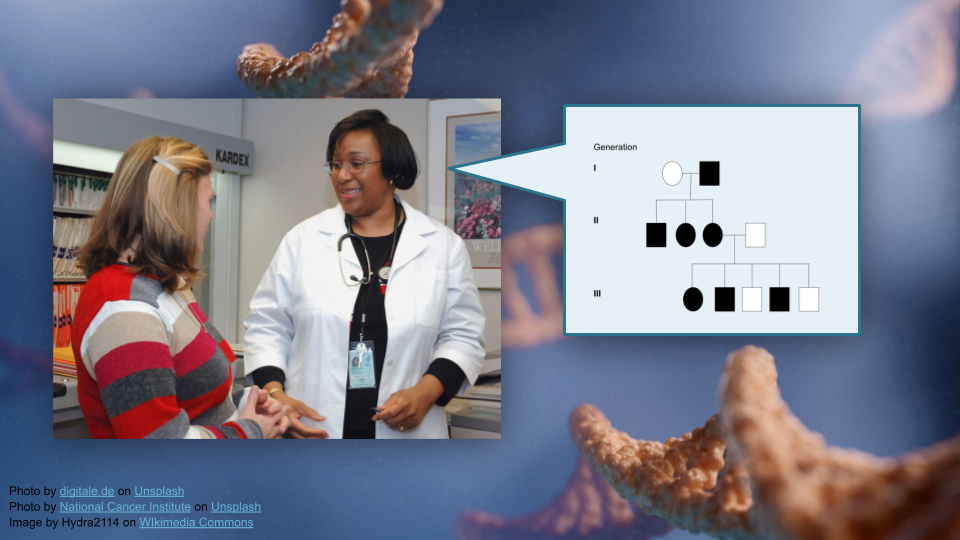
This activity teaches students to interpret pedigree information and use bioinformatics tools (R) to build pedigrees for tracking disease traits in families. Students will work with real data from a bipolar disorder genetics study to understand how researchers understand risk genes using pedigrees.
Duration: Approximately 2-3 hours (depending on student familiarity with R) (help us improve this estimate!)
Learning Objectives
Interpret pedigree information to determine the suitability of a DNA marker for tracking a disease trait in a family (see GSA learning objectives).
Build a pedigree using bioinformatics tools (R).
Prerequisites
Students will benefit from some awareness of genetic inheritance models, pedigrees, and genetically-based medical conditions. We suggest this activity could accompany lectures that discuss these topics.
Evaluation
GEMs is an NIH-funded program. Part of our mission is understanding the impact of our materials. Please take the time to review our program as an instructor after this activity. We also appreciate you distributing our survey to students before and after they participate in GEMs content.
You can view our IRB approval here. Feel free to contact the GEMs team with any questions (gems at fredhutch dot org).
Materials
Student Activity
You can use this module in several formats. Feel free to adapt to your needs!
Students will need RStudio, data, and the R package: kinship2. This means either:
- A device with an internet connection for this activity as written (using Posit Cloud), access to the data hosted at https://genomicseducation.org/data/pedigree_data.csv.
- A local installation of R or RStudio, the ability to install the
kinship2package, and downloaded data from https://genomicseducation.org/data/pedigree_data.csv. Instructors are welcome to download the data and provide it via email or an LMS system if desired.
Instructor Materials
A Google Slides presentation is available for borrowing images here.
An answer key is available here. Please message Ava Hoffman (ahoffma2 at fredhutch dot org) to get access.
Scientific Topics
The activity is based a published research study “Rare variants implicate NMDA receptor signaling and cerebellar gene networks in risk for bipolar disorder.” Students will gain exposure to the following:
Bipolar Disorder: Neuropsychiatric condition affecting ~1% of population, with 10-25% risk for children of affected parents
Cerebellum: Brain region containing 50% of neurons despite being 10% of brain volume; involved in motor coordination and emotional regulation
NMDA Receptors: Glutamate receptors crucial for memory and learning; blocked by substances like alcohol and ketamine
DAO Gene: D-amino acid oxidase gene identified as significantly associated with bipolar disorder risk
Outline
Part 1: Background and Setup (30-45 minutes)
Part 2: Basic Pedigree Construction (20-30 minutes)
Part 3: Adding Diagnostic Information (15-20 minutes)
Part 4: Genotype Mapping and Analysis (30-40 minutes)