Database Detectives: Exploring Public Genomic Databases Student Guide
You are a genetic detective! Your lab group has been running experiments for a variety of Drosophila genes. Unfortunately, there was an accident with a broken pipe and the computer, and all the stored data is gone! You have copies of the experimental results, but you haven’t been able to find the key that tells you which set of experiments corresponds to which gene. Your job is to find the background information on each gene using online databases, then match the fruit fly gene to the correct set of experimental results.
The suspected genes
Each mystery experiment is associated with one of four Drosophila genes: scarlet, plum, mustard, and white.
Each of these genes has been characterized in the Drosophila genome and you know their common name, but not much else. You’ll need to figure out where they’re located in the Drosophila genome, in what tissues they’re expressed, and what species share homologous genes. Finally, you’ll need to track down what possible human diseases their human homologs might be involved in.
The databases
For this activity, you’ll explore a variety of curated genomic databases. Many of these databases are available to the public at little or no cost and are supported by grants to the researchers and organizations that maintain them.
Curated databases are simply databases where information is screened, validated, and organized by experts to ensure the data are accurate and reliable. Information has undergone rigorous quality control checks and includes sources and contextual information to help users understand the data. The data is generally standardized and organized in a way to be easily accessible and searchable.
It’s important to remember that the databases in this activity aren’t the only databases that exist! There are many more databases out there, each with its own focus. Luckily, most databases are organized in similar ways, so once you learn how to navigate one database, you’ll be able to easily figure out how to navigate new ones.
You should also keep in mind that some information can be found in multiple databases. For example, many databases that focus on the genome of a particular model organism will also have information about the human diseases associated with that particular gene. This information can also be found in databases that focus specifically on human genetic diseases (like OMIM or MalaCards). Just like there are many ways to cook an egg, there’s many ways to find the genomic information you might need in public databases. As long as you are careful and using a curated database, you can use the one you’re most comfortable with.
Part 1
Start with exploring FlyBase to get some basic information about each of the four genes.
FlyBase is a database focused on Drosophila genetics, genomics, and functional data. It is maintained by a consortium of Drosophila researchers and computer scientists at Harvard University, University of Cambridge (UK), Indiana University, and the University of New Mexico [1].
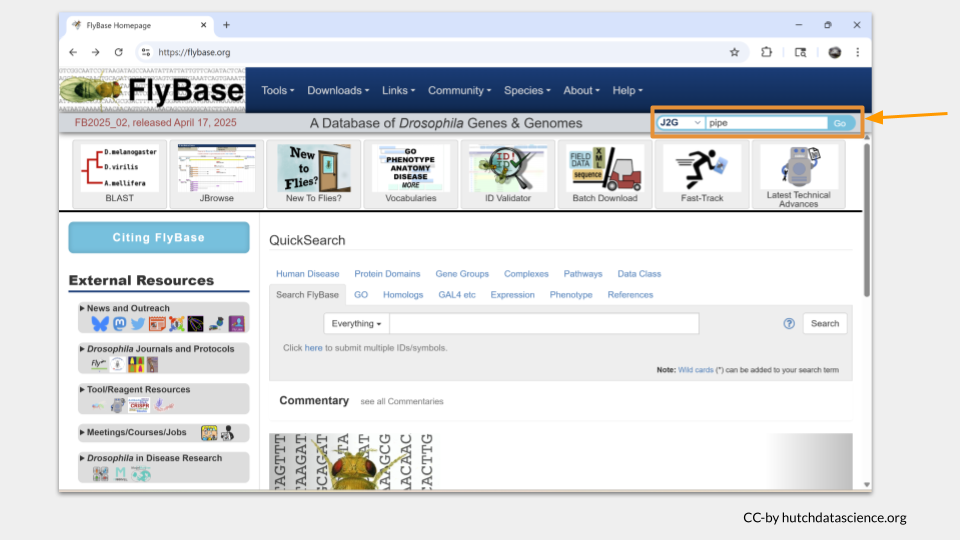
First, open the FlyBase website by clicking the link or typing “https://flybase.org” into your browser. Then, type the name of the gene into the “J2G” bar in the upper right hand corner. (J2G stands for “Jump to Gene”.) As an example, try typing the gene name pipe and click enter.
The gene page for pipe will load. On the top of the page, you will see the general information about this gene, including the FlyBase gene ID and a brief description of what this gene does.
Below this general information section, the genomic location data will appear. There are several different ways gene locations might be recorded; for our purposes, we’re interested in the sequence location. The pipe gene is located. “3L” means this gene is on the left arm of the third chromosome. The numbers after 3L refer to the nucleotide location.
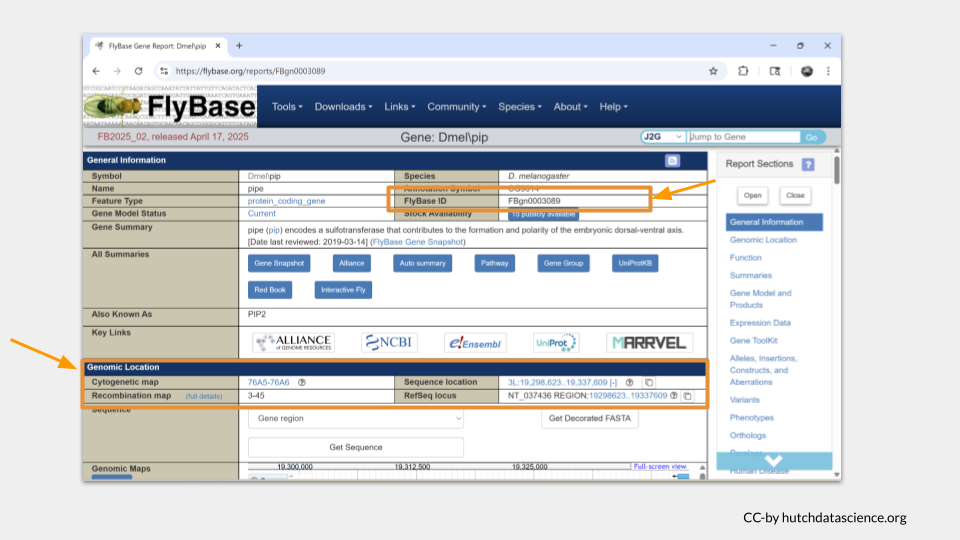
You can scroll down the page or click the menu on the right-hand side to move to additional sections. The next section we’re interested in is the function section. Here, we can see what the general molecular function of the gene is, what sorts of biological processes the gene product (the protein) is involved in, and where the protein is found in the cell, or the cellular component. In the case of pipe, we can see that it’s an enzyme involved in development, gene expression, protein metabolism. We also see that the pipe protein is found in the membrane and endomembrane system, as well as in the nucleus around the chromosomes.
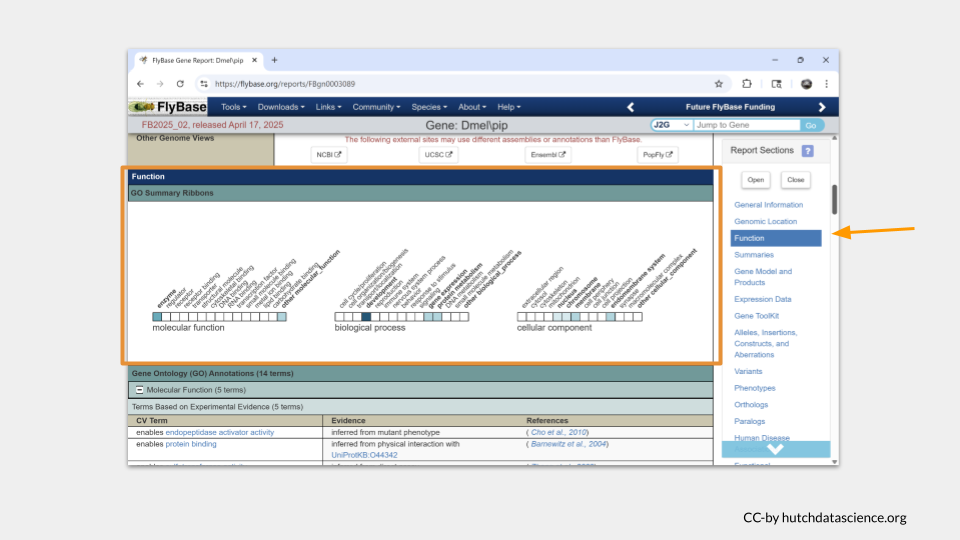
Scrolling down further takes us to a section with more details about the gene function. You may need to click on the small plus sign to view the details (or click on the minus sign to hide the details). We can see that pipe codes for an enzyme that is enables endopeptidase activator activity and protein binding.
Now it’s your turn to look up your four genes in FlyBase.
For each gene (scarlet, plum, mustard, and white), find the following information:
- FlyBase gene ID
- chromosomal location
- molecular function
- the biological processes the gene product is involved in
- where in the cell the gene product can be found
Part 2
Next, let’s explore additional genomic databases beyond FlyBase to characterize the homologs and orthologs to the four Drosophila genes.
A homolog is a gene or protein that is similar to another due to having a common evolutionary origin. Many genes in Drosophila have homologs in the human genome, as well as in the genomes of common model organisms like mice and zebrafish.
An ortholog is another term for a gene or protein that is similar due to having a common evolutionary origin, but in order to be an ortholog, the two genes must also have similar functions within the different species. An ortholog is a specialized kind of homolog.
Genomic databases will report both homologs and orthologs.
First, we need to find out what the name of the homologous genes are. The gene IDs and common names differ between species. If we keep scrolling down the page on the pipe gene in FlyBase, we can find what those homologous genes are called in our species of interest.
It turns out that the human gene most likely to be a homolog of pipe is called UST.
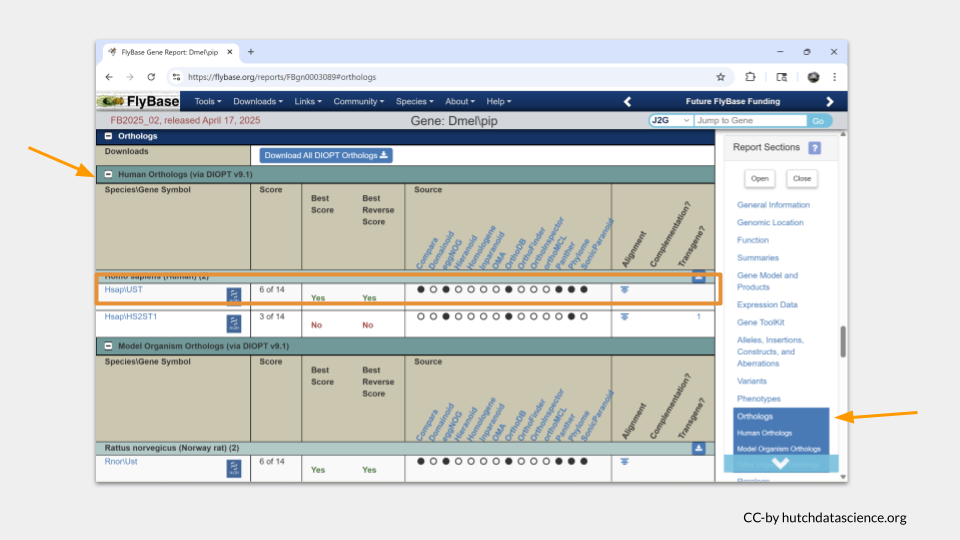
If we scroll down a little farther, we see that the most likely homologous mouse (Mus musculus) is called Ust. In the zebrafish (Danio rerio), it is most likely to be usta.
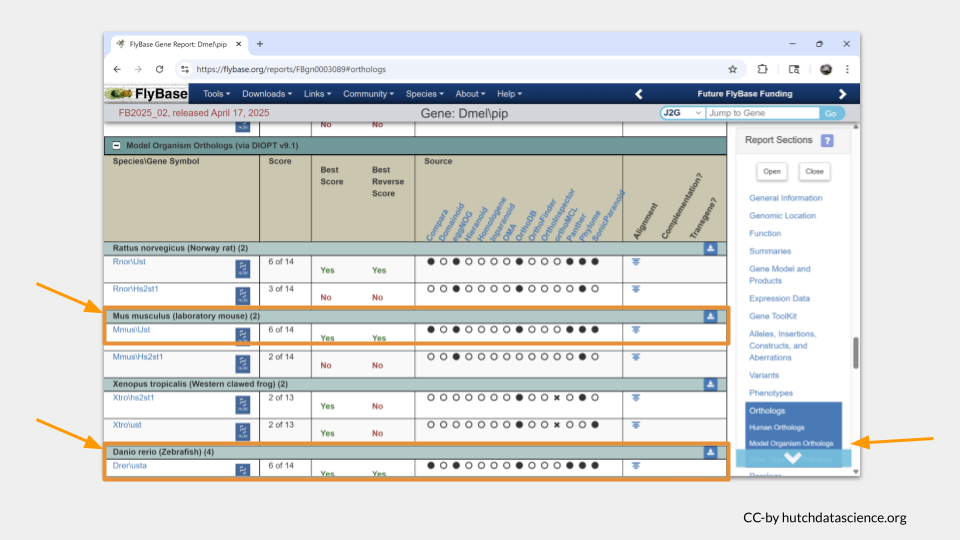
To find out more information about these homologs, we’re going to need to look at additional species-specific databases. Let’s start with the mouse.
The Mouse Genome Informatics database is the international database for the laboratory mouse, integrating genetic, genomic, and biological data as part of an effort to use the mouse to study human health and disease. It is maintained by the Jackson Laboratory [2].
Why do we have a database devoted to the mouse?
The mouse is the most commonly-used model organism in laboratory work. In fact, mice and rats make up 95% of the lab animal population, and more than 80% of the research that has been awarded the Nobel Prize for Medicine was done at least in part with mouse models (https://www.cshl.edu/of-mice-and-model-organisms/, https://fbresearch.org/medical-advances/nobel-prizes).
So what makes mice such good model organisms for biomedical research? One reason is that mice and humans are both mammals and have about 85% of their protein-coding genome in common. As a result, mouse physiology is quite similar to human physiology. The mouse circulatory, reproductive, digestive, hormonal, and nervous systems are frequently used as models to study how humans grow, age, and develop chronic diseases.
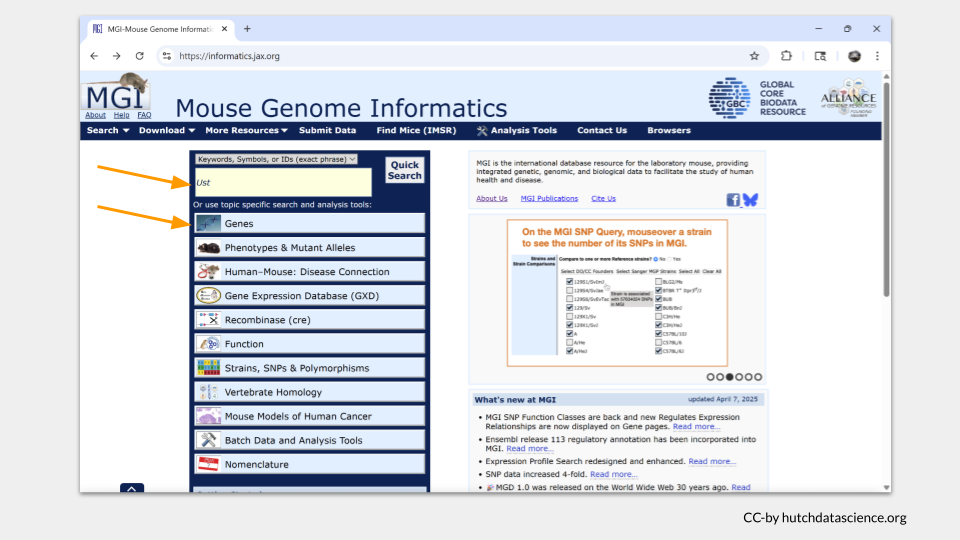
Open the MGI database and search for the name of the mouse homolog Ust. You can do this either by typing Ust into the search bar and clicking “Quick Search” or by doing a topic specific search with the “Genes” toolbar.
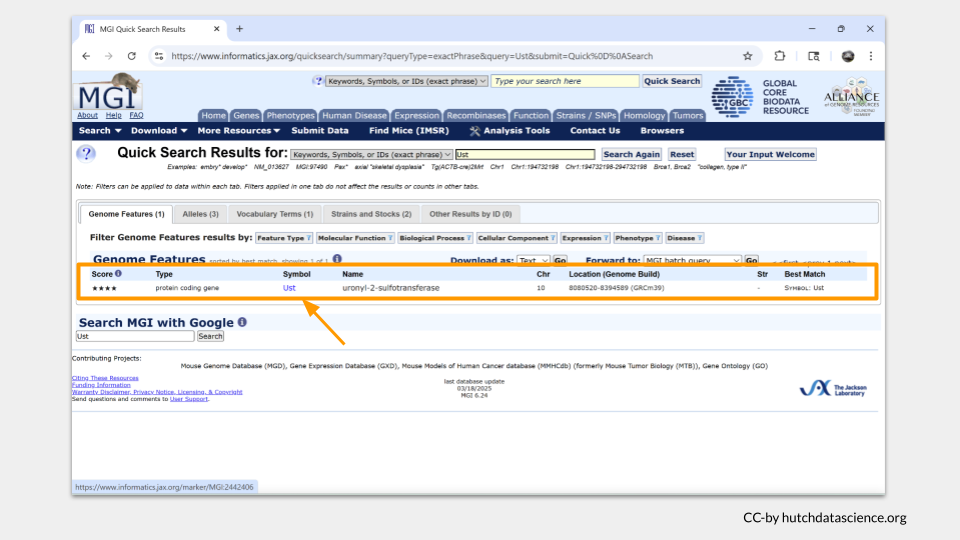
A “Genome Features” summary table should open up with information about Ust. MGI tells us that Ust is a protein-coding gene on chromosome 10. Clicking on the highlighted gene symbol will take us to a page with more detail.
The kinds of information you found in FlyBase will also be present on the detailed gene page in MGI. The information is organized into sections, similar to what you’ve already experienced with FlyBase. The fourth section down includes information about the human ortholog to Ust - it turns out to be UST, which we already knew from FlyBase.
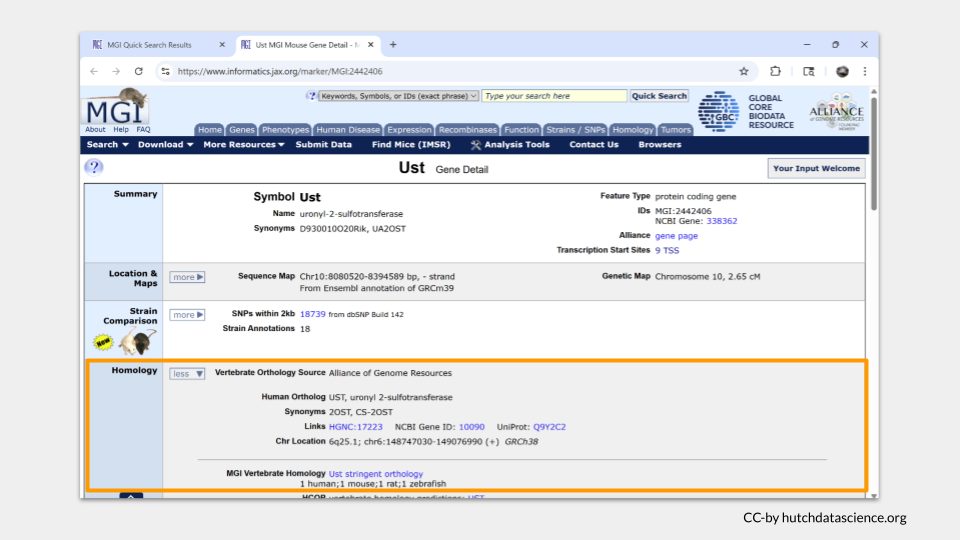
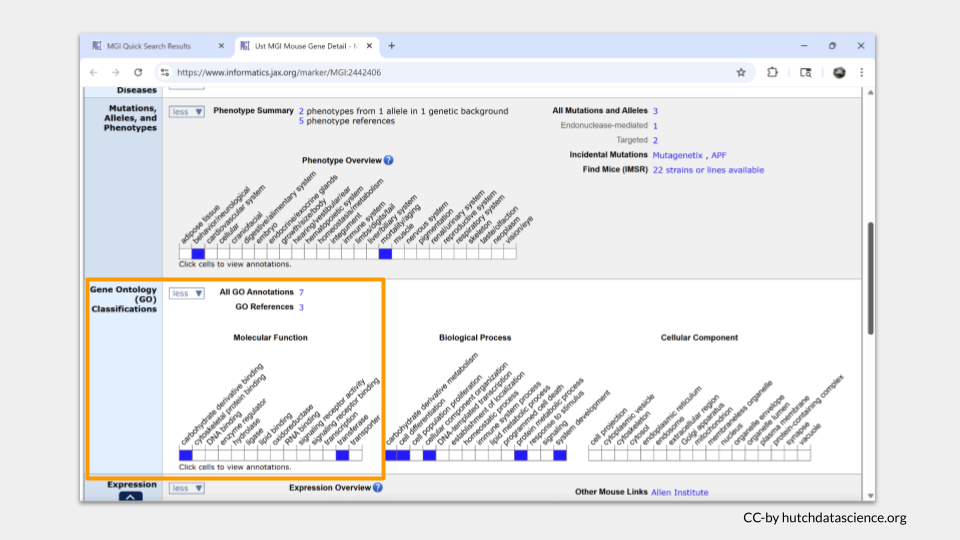
Farther down the page is a section called “Gene Ontology (GO) Classifications”. This is the section that has information about the molecular functions, biological processes, and cellular components for Ust.
“Gene ontology” is a bioinformatics initiative focused on standardizing the vocabulary and annotations researchers use to describe genes, proteins, and data about gene and protein function. This effort allows us to more easily make comparisons about homologous and orthologous genes and proteins across species.
In mice, Ust is a tranferase (an enzyme!) involved in carbohydrate derivative binding.
In order to look up information about the zebrafish homolog, we need to switch databases again.
The ZFIN (Zebrafish Information Network is the database of genetic and genomic data for the zebrafish as a model organism. It was formed by a group of zebrafish researchers at Cold Spring Harbor and is maintained by the ZFIN Database Team at the University of Oregon [3].
Why do we have a database devoted to the zebrafish?
Zebrafish are a popular model organism for researchers who focus on developmental biology, genetics, and modeling diseases. They develop rapidly and have high fertility, so it can be relatively quick and fast to study the effects of genes or diseases over multiple generations. They are also transparent, which is especially useful for watching how organs develop as the zebrafish matures.
Zebrafish share a high degree of genetic homology with humans!
Open ZFIN and search for the zebrafish homolog usta. The ZFIN search bar will offer some options to refine your search. Choose “Gene/Transcript” from the drop down menu.
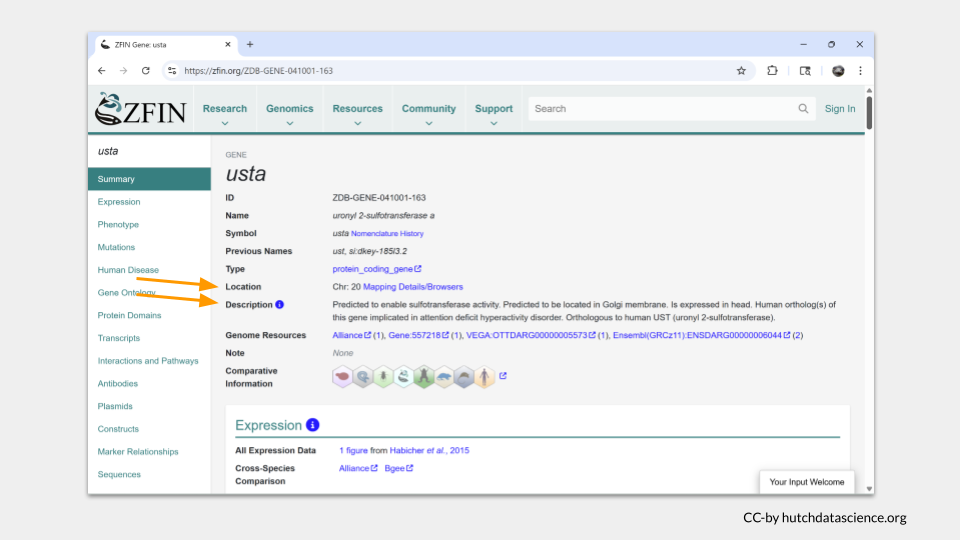
ZFIN takes you directly to the gene page for usta. A summary of basic information about usta is at the top, with more detailed sections as you scroll down. Without much searching, we discover that usta is located on chromosome 20 and is predicted to enable sulfotransferase activity.
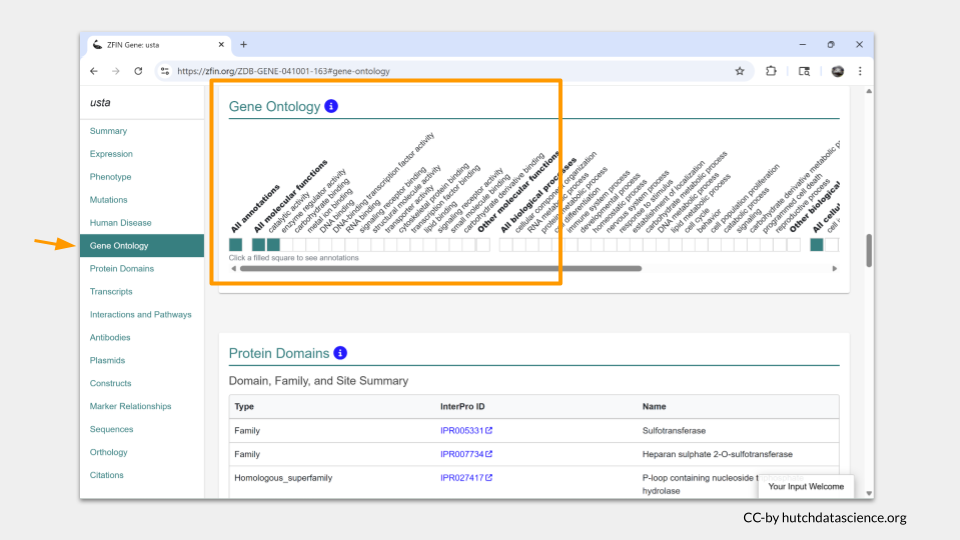
Clicking on “Gene Ontology” in the menu on the left takes you to a more detailed list of the molecular functions associated with usta. The summary on top just mentioned the words “sulfotransferase activity”; in the Gene Ontology section, we learn that usta is involved in catalytic activity and enzyme regulator activity as well.
Finally, we can turn to the human homolog UST. There are several databases with information about human genes. GeneCards is a user-friendly database that collects and displays information about human genes from multiple research databases. It is maintained as a joint effort between the Weizmann Institute of Science and LifeMap Sciences [4].
Open the GeneCards page and search for the human homolog UST in the “Explore a Gene” search bar.
Unlike MGI or ZFIN, the GeneCards page on our searched gene doesn’t have the information about location at the top. Instead, we will need to click on “Genomics” in the header menu.
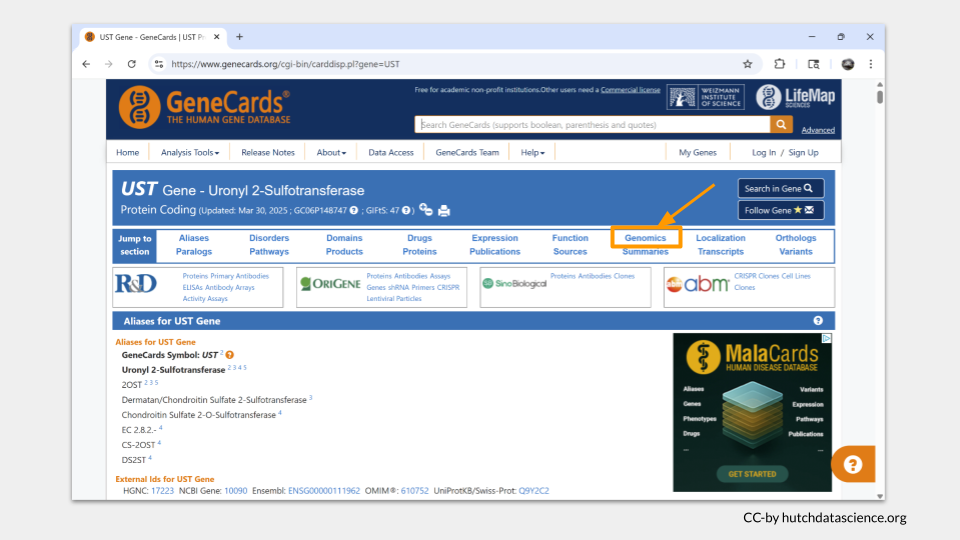
The genomics section of the GeneCards page includes location information for multiple assemblies, as well as a map of the chromosome. UST is located on chromosome 6.
A genome assembly is just a version of the genome map. As scientists unlock more detail about genomes, they periodically publish an updated “reference genome”, with everything we have discovered about where genes, enhancers, promotors, and non-coding regions are located in the genome. Think of an updated genome assembly as a better map.
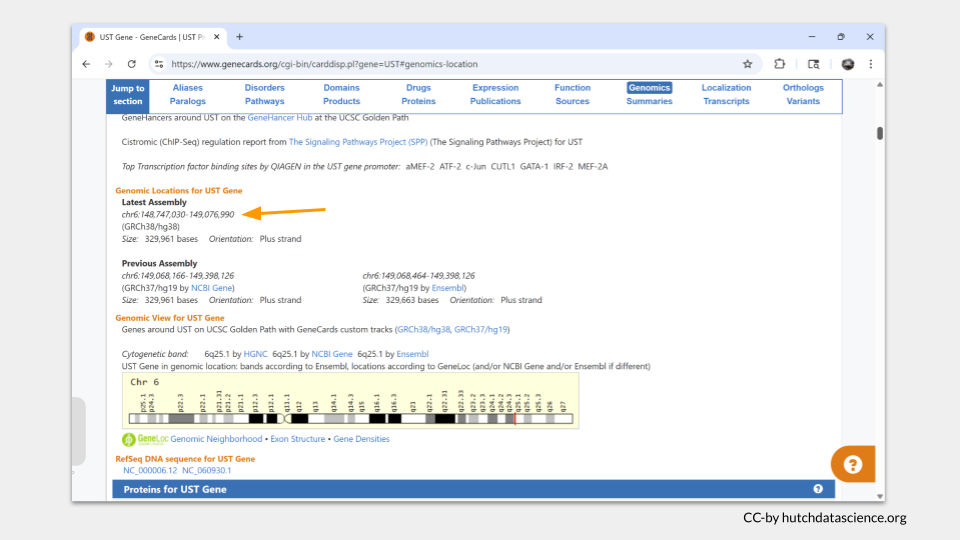
We can also find information about UST’s molecular function. Choosing “Function” from the menu takes us to a list of gene ontology terms (commonly called GO terms) associated with UST. There are a lot of words you might recognize, including sulfotransferase activity.
Now it’s your turn to look up information about the homologs of your four Drosophila genes.
For each of the Drosophila genes (scarlet, plum, mustard, and white), find the following information for the mouse, zebrafish, and human homologs:
- name
- chromosomal location
- molecular function
Does this information differ among the homologs? For example, do the homologs of scarlet do the same things as scarlet?
Part 3
In addition to the experimental data you’re sorting through, you notice that someone has collected literature on different human diseases. You assume these diseases are thought to be caused in part by some of the human homologs of the genes your lab is studying, so you decide to look up the information using a pair of human disease databases.
OMIM, short for Online Mendelian Inheritance in Man, is a comprehensive database of human genes and genetic phenotypes. It is maintained by the McKusick-Nathans Institute of Genetic Medicine at Johns Hopkins University.
MalaCards is a searchable database offering information on human diseases, medical conditions, and disorders. It is maintained as a joint effort between the Weizmann Institute of Science and LifeMap Sciences.
Both databases will give you the information you need. We’ll go through how to use both, but you only need to use one for this activity.
Let’s start with OMIM. Click on the link or type “https://omim.org” into your browser. On the landing page, you can type either a disease or a gene name into the search bar. (Just remember to use the human gene name or gene ID!)
Earlier, we discovered the human homolog of pipe is UST. Let’s look this gene up and see what (if any) diseases it might be associated with.
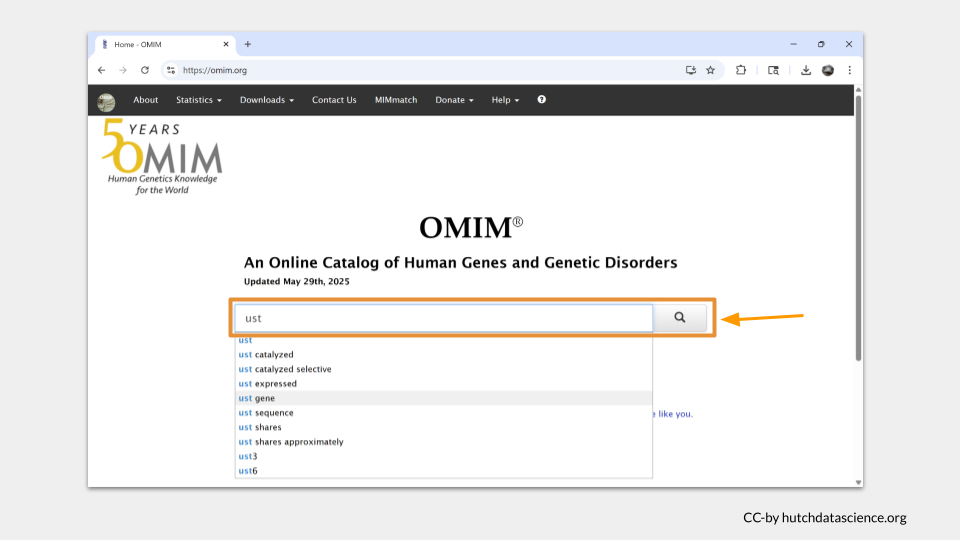
Our search pulls up a list of entries associated with our search term. For UST, there’s only one. Click on it.
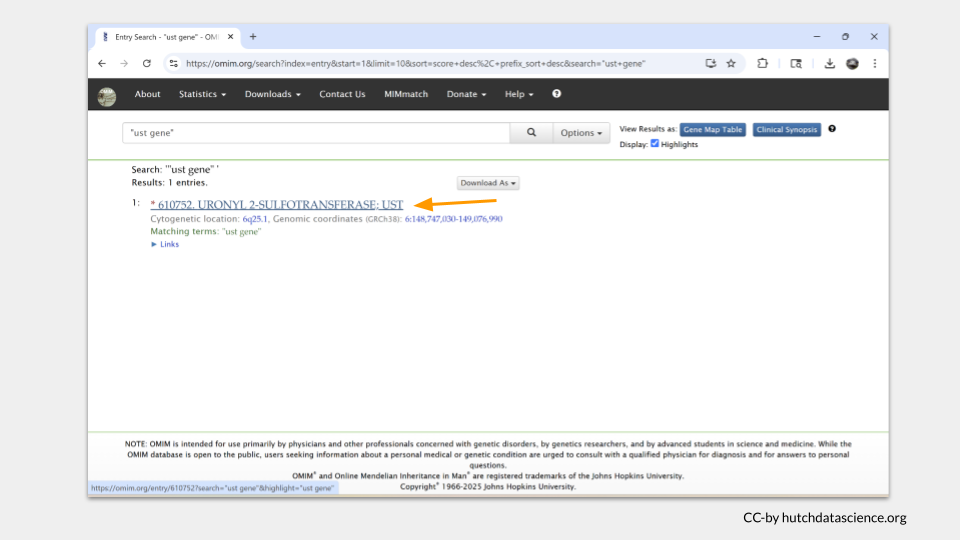
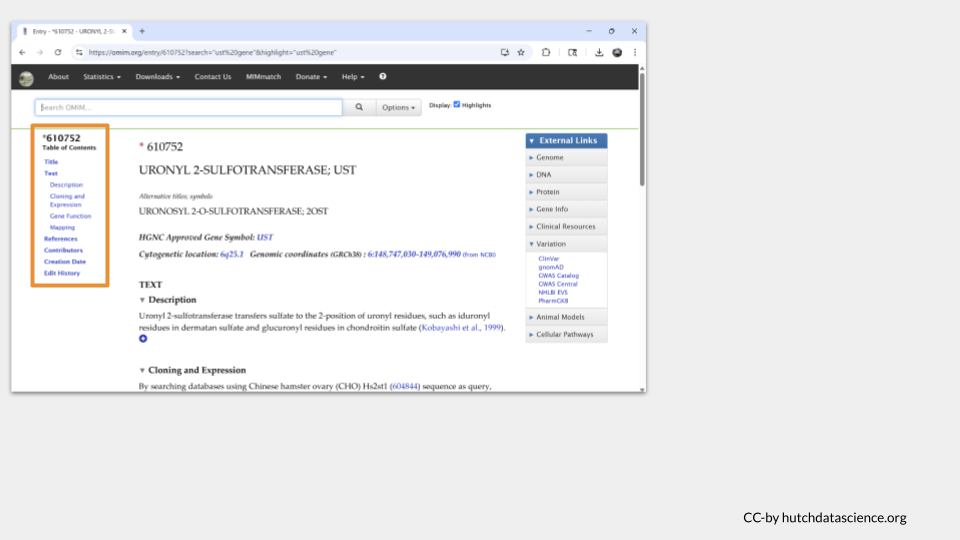
Each entry in OMIM will have a table of contents on the left hand side of the page. In the case of UST, there’s not much there. In fact, there doesn’t seem to be any subsection focusing on disease state or phenotype.
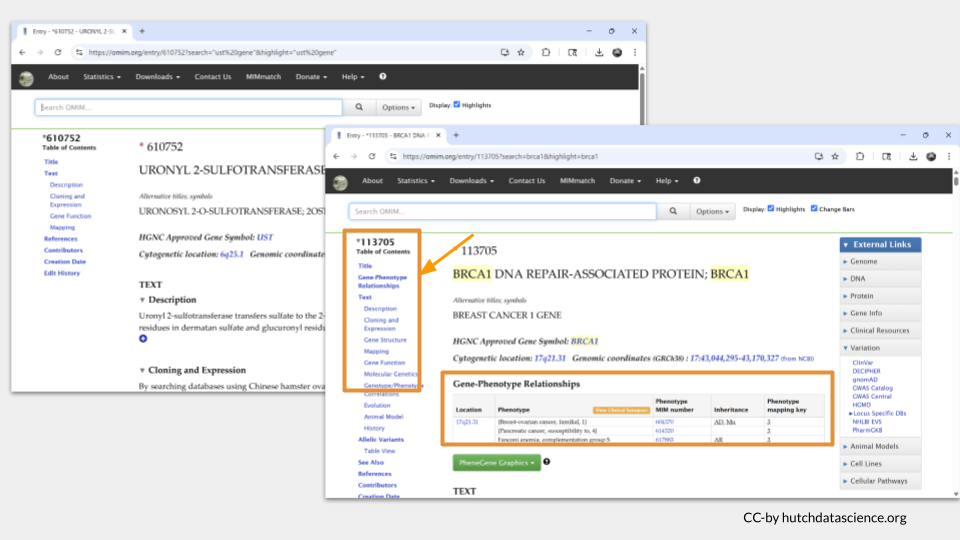
As a comparison, we can search for the known disease risk gene BRCA1, which is associated with an increased risk of breast and ovarian cancer. When we look at the top OMIM hit for BRCA1, we can see a section in the table of contents called “Gene-Phenotype Relationships”. This is the section where OMIM displays any possible disease phenotype associated with the gene.
Since we don’t see this section for UST, we can conclude that UST doesn’t appear to cause any human diseases that we know of at this time.
You can find similar information on the MalaCards website. Click the link or type “https://www.malacards.org” into your browser to open the webpage. Then type “UST” into the bottom search bar. (Although you can look up genes on MalaCards, most of the time you look up a a disease to find out what genes are associated with it instead of the other way around.)
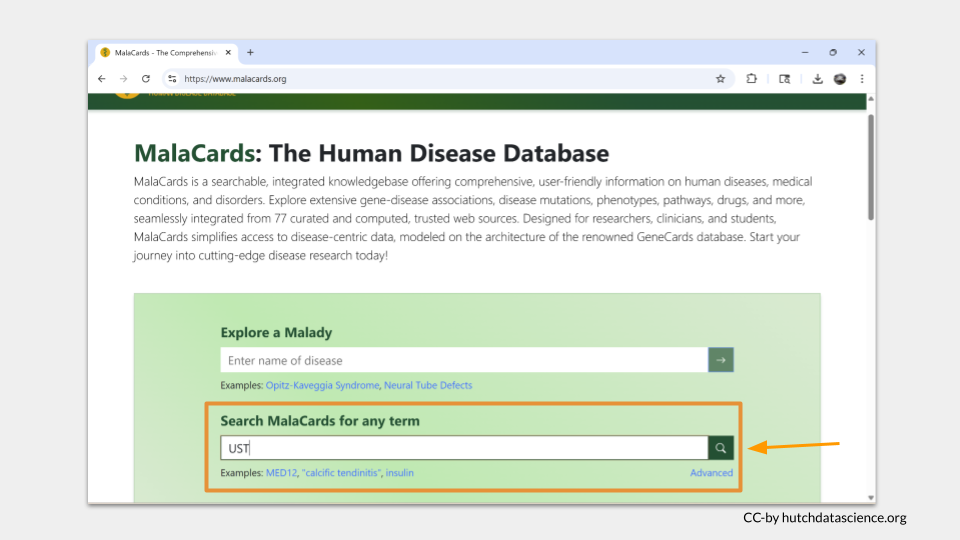
Since we used the “search any term” search bar, MalaCards will look for the term “UST” on all the disease pages in the database. The results are ordered with the best-matching page on top. Click “Crohn’s Disease” to see why “UST” matched this entry.
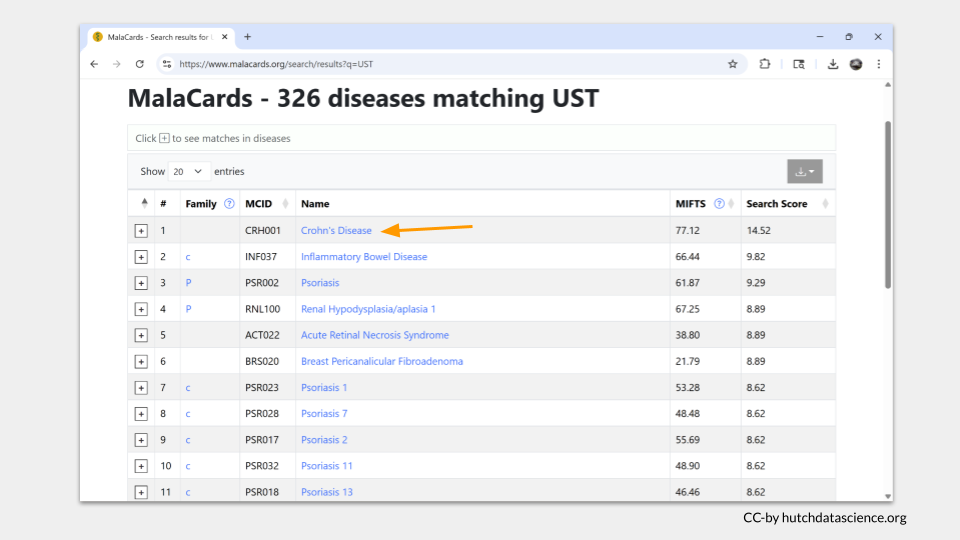
One nice thing about MalaCards is that the search term is highlighted in yellow when you open the page. In this case, we can see that “UST” actually matched part of drug that was investigated as a treatment for Crohn’s disease. This suggests that the gene UST is not found in any of the MalaCards disease entry.
Let’s search MalaCards with the term “BRCA1” to see what it looks like when the gene name is found in an entry. In the top hit entry (“Hereditary Breast Ovarian Cancer Syndrome), we can see the actual gene name BRCA1 shows up in the overview, as well as in the section about associated genes and the alternate name for the disease.
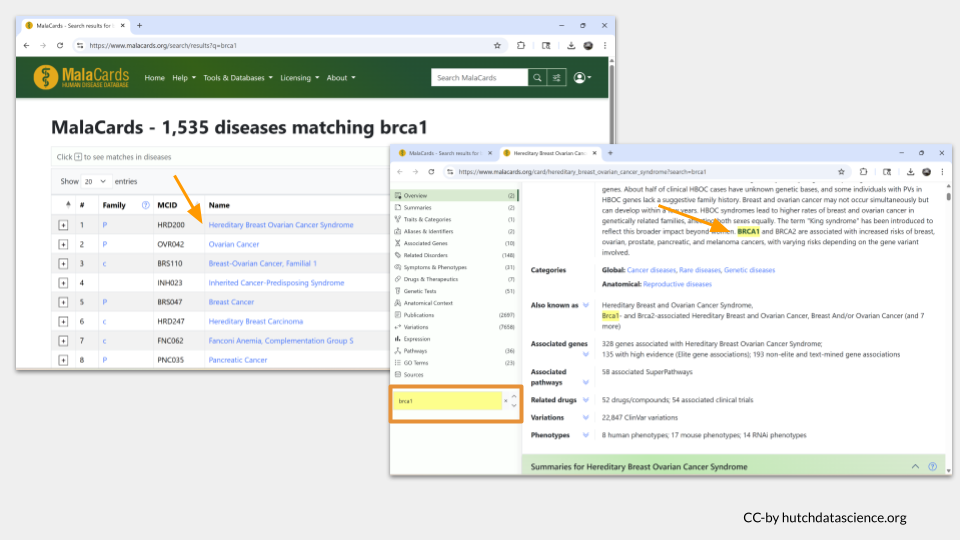
Now it’s your turn to look up information the diseases (if any) your four human homologs might be associated with.
What human disease or disorder (if any) are the homologs of scarlet, plum, mustard, and white associated with?
Part 4
Finally, it’s time for you to report back to your supervisor. Here’s what you know about the experiments:
Experiment 1: the data seems to be from the 3R chromosome in Drosophila and measures whether motor neurons are being pruned at the same rate in the experimental group (versus the control group)
Experiment 2: this data includes measurements on transmembrane transport in the zebrafish, specifically in intestinal cells
Experiment 3: this data looks at mouse cells in culture and tracks the damage caused by exposure to reactive oxygen species (ROS) and oxidative stress
Experiment 4: this data includes sequences from the Drosophila X chromosome and appears to be a genome annotation project
There is also extensive background research about cerebellar atrophy, gallbladder disease, and hyperuricemia (high uric acid concentration in the urine). You also find some initial research looking at neuronal cell adhesion.
Which Drosophila genes are associated with each experiment?
Which background research on human disease belongs with each experiment?